NOT Sharding RavenDB Vector Search
I ran into an interesting post, "Sharding Pgvector," in which PgDog (provider of scaling solutions for Postgres) discusses scaling pgvector indexes (HNSW and IVFFlat) across multiple machines to manage large-scale embeddings efficiently. This approach speeds up searches and improves recall by distributing vector data, addressing the limitations of fitting large indexes into memory on a single machine.
That was interesting to me because they specifically mention this Wikipedia dataset, consisting of 35.1 million vectors. That… is not really enough to justify sharding, in my eyes. The dataset is about 120GB of Parquet files, so I threw that into RavenDB using the following format:
Each vector has 768 dimensions in this dataset.
33 minutes later, I had the full dataset in RavenDB, taking 163 GB of storage space. The next step was to define a vector search index, like so:
from a in docs.Articles
select new
{
Vector = CreateVector(a.Embedding)
}That index (using the HNSW algorithm) is all that is required to start doing proper vector searches in RavenDB.
Here is what this looks like - we have 163GB for the raw data, and the index itself is 119 GB.
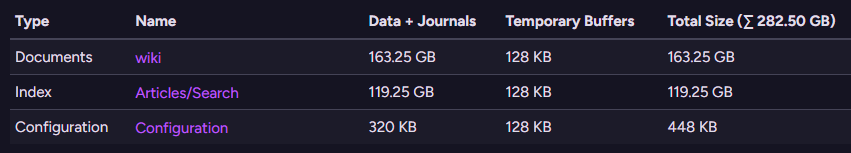
RavenDB (and PgVector) actually need to store the vectors twice - once in the data itself and once in the index.
Given the size of the dataset, I used a machine with 192 GB of RAM to create the index. Note that this still means the total data size is about ⅓ bigger than the available memory, meaning we cannot compute it all in memory.
This deserves a proper explanation - HNSW is a graph algorithm that assumes you can cheaply access any part of the graph during the indexing process. Indeed, this is effectively doing pure random reads on the entire dataset. You would generally run this on a machine with at least 192 GB of RAM. I assume this is why pgvector required sharding for this dataset.
I decided to test it out on several different machines. The key aspect here is the size of memory, I’m ignoring CPU counts and type, they aren’t the bottleneck for this scenario. As a reminder, we are talking about a total data size that is close to 300 GB.
| RAM | RavenDB indexing time: |
| 192 GB | 2 hours, 20 minutes |
| 64 GB | 14 hours, 8 minutes |
| 32 GB | 37 hours, 40 minutes |
Note that all of those were run on a single machine, all using local NVMe disk. And yes, that is less than two days to index that much data on a machine that is grossly inadequate for it.
I should note that on the smaller machines, query times are typically ~5ms, so even with a lot of data indexed, the actual search doesn’t need to run on a machine with a lot of memory.
In short, I don’t see a reason why you would need to use sharding for that amount of data. It can comfortably fit inside a reasonably sized machine, with room to spare.
I should also note that the original post talks about using the IVFFlat algorithm instead of HNSW. Pgvector supports both, but RavenDB only uses HNSW. From a technical perspective, I would love to be able to use IVFFlat, since it is a much more traditional algorithm for databases.
You run k-means over your data to find the centroids (so you can split the data into reasonably sized chunks), and then just do an efficient linear search on that small chunk as needed. It fits much more nicely into the way databases typically work. However, it also has some significant drawbacks:
- You have to have the data upfront, you cannot build it incrementally.
- The effectiveness of the IVFFlat index degrades over time with inserts & deletes, because the original centroids are no longer as accurate.
Because of those reasons, we didn’t implement that. HNSW is a far more complex algorithm, both in terms of the actual approach and the number of hoops we had to go through to implement that efficiently, but as you can see, it is able to provide good results even on large datasets, can be built incrementally and doesn’t degrade over time.
Head-to-head comparison
I decided to run pgvector and RavenDB on the same dataset to get some concrete ideas about their performance. Because I didn’t feel like waiting for hours, I decided to use this dataset. It has 485,859 vectors and about 1.6 GB of data.
RavenDB indexed that in 1 minute and 17 seconds. My first attempt with pgvector took over 7 minutes when setting maintenance_work_mem = '512MB'. I had to increase it to 2GB to get more reasonable results (and then it was 1 minute and 49 seconds).
RavenDB is able to handle it a lot better when there isn’t enough memory to keep it all in RAM, while pgvector seems to degrade badly.
Summary
In short, I don’t think that you should need to go for sharding (and its associated complexity) for that amount of data. And I say that as someone whose database has native sharding capabilities.
For best performance, you should run large vector indexes on machines with plenty of RAM, but even without that, RavenDB does an okay job of keeping things ticking.






Comments
Comment preview